scquill 0.3.2
pip install scquill
Latest version
Released:
Approximate any single cell data set, saving >99% of memory and runtime.
Navigation
Unverified details
These details have not been verified by PyPIMeta
- License: MIT License (MIT)
- Author: Fabio Zanini
- Requires: Python <3.13, >=3.11
Classifiers
- License
- Programming Language
Project description
scquill
Approximate any single cell data set, saving >99% of memory and runtime.
It's pronounced /ˈskwɪɹl̩//, like the animal.
Approximating a single cell data set
import scquill
q = scquill.Compressor(
filename='myscdata.h5ad',
output_filename='myapprox.h5',
celltype_column="cell_annotation",
)
q()
Exploring an approximation
To load an approximation:
import scquill
app = scquill.Approximation(
filename='myapprox.h5',
)
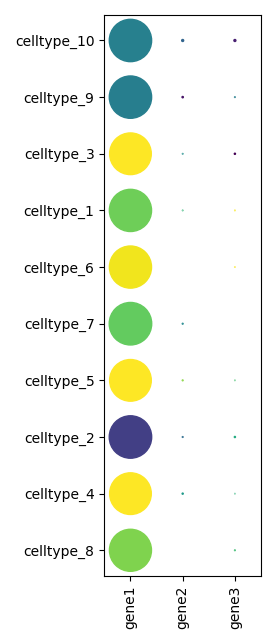
To show a dot plot:
scquill.pl.dotplot(app, ['gene1', 'gene2', 'gene3'])
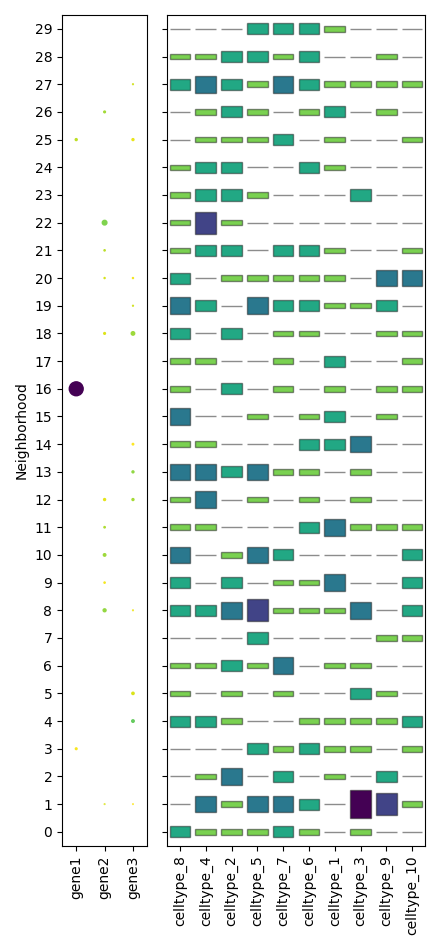
To show a neighborhood plot:
scquill.pl.neighborhoodplot(app, ['gene1', 'gene2', 'gene3'])
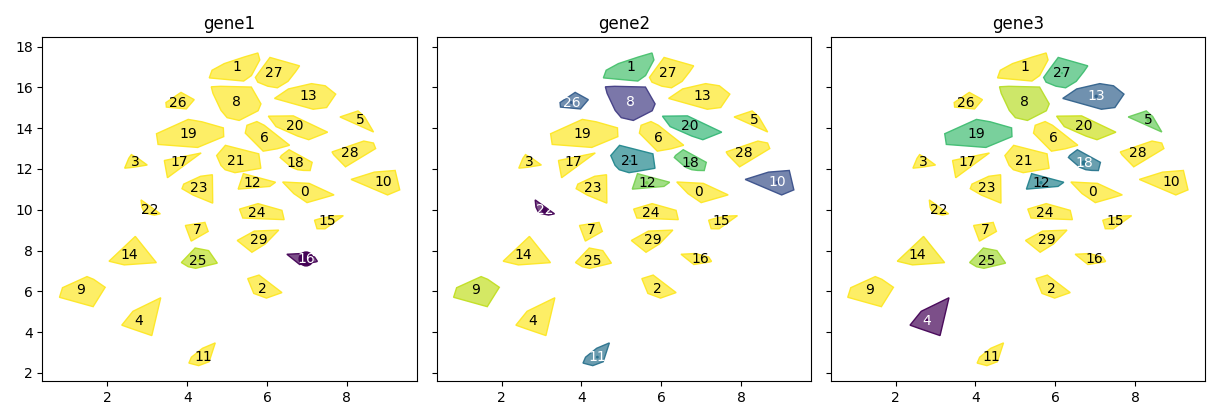
To show embeddings of cell neighborhoods, similar to single-cell UMAPs:
scquill.pl.embedding(app, ['gene1', 'gene2', 'gene3'])
MORE TO COME
Authors
Fabio Zanini @fabilab
Project details
Unverified details
These details have not been verified by PyPIMeta
- License: MIT License (MIT)
- Author: Fabio Zanini
- Requires: Python <3.13, >=3.11
Classifiers
- License
- Programming Language
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file scquill-0.3.2.tar.gz.
File metadata
- Download URL: scquill-0.3.2.tar.gz
- Upload date:
- Size: 19.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.3 CPython/3.12.6 Linux/6.6.23-1-lts
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b4212a2be9a39c84a64bd4dbdc53ac492acd0c2f839f7c4333ae90f52b67237b |
|
| MD5 | f7da46ad1e811f473d4b3fda1e4254e7 |
|
| BLAKE2b-256 | 4fe2d4cd0baf39e2c6a1df2743014ad0c0b0368ac910206c896bddb263a42e3f |
File details
Details for the file scquill-0.3.2-py3-none-any.whl.
File metadata
- Download URL: scquill-0.3.2-py3-none-any.whl
- Upload date:
- Size: 26.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.3 CPython/3.12.6 Linux/6.6.23-1-lts
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ce310266fe7bc80460faaa48b3cecf2ad2a67979366f8703c6b444022c99a52e |
|
| MD5 | 850e683dfd8525cda8053ced88f1e4fa |
|
| BLAKE2b-256 | 921918354d299523c86c633618127f961cf8add240aaad06eb28be7cb2c33e1e |











