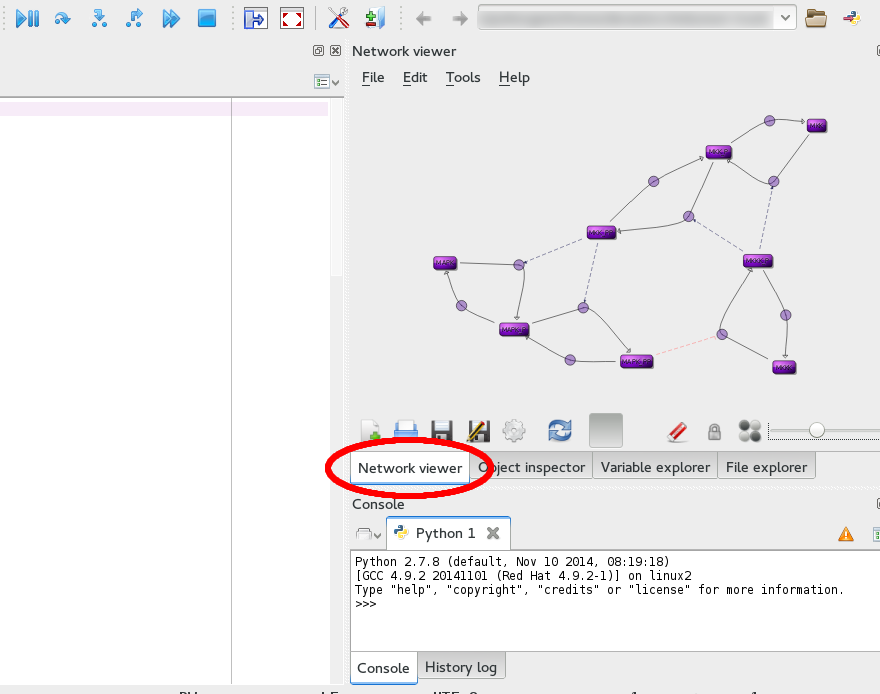
libSBNW is a network viewer which supports autolayout of reaction networks and uses libSBML for reading/writing models. The intent of this project is to provide user and developers of systems/synthetic biology software tools a convenient, portable library to enable automatic layout of SBML models and to record this information in the SBML layout extension. To facilitate this, the functionality of the library is provided as a custom plugin within the Spyder-based Tellurium modeling environment. For developers, pre-built binaries including bindings for Python 2 are provided via the release page on this site.
The library is designed as a reusable implementation of an automatic layout for SBML. As such, it has many downstream uses. To name a few:
- Called by Python code via the included Python bindings
- Via the network viewer GUI plugin for Tellurium
- Directly via the C API in a pre-existing project
- As part of e.g. a Django web service
The network viewer plugin is part of Tellurium, which can be downloaded here for Windows and Mac OS X. Separate binaries for the libSBNW library are available from SourceForge.
Video tutorials are available here which demonstrate how to use the network viewer plugin in Tellurium.
The SBML test models used in development of this tool are available for download from the releases page as well. The network viewer is frequently tested with these files.
Online C API documentation (via Doxygen) can be found at http://sys-bio.github.io/sbnw/docs. Python documentation can be found at http://sys-bio.github.io/sbnw/sphinx.
- On Mac, extract the contents of http://sourceforge.net/projects/sbnw/files/1.2.7/sbnw-1.2.7-os-x-site-packages-py2.tar.bz2/download to your site-packages directory.
- On Windows, extract the contents of http://sourceforge.net/projects/sbnw/files/1.2.7/sbnw-1.2.7-site-packages-win32.zip/download to your site-packages directory.
-
Install the latest version of libSBML or build it from source (tested with 5.6, 5.8, 5.10, 5.11). (VIDEO showing steps to follow to build from source using CMAKE and Visual Studio: https://www.youtube.com/watch?v=e_Lydwzx-Hg, note that a few steps differ from what is shown in the video: specifically,
- you can download the 64-bit versions if you like; in the video the 32-bit versions are used
- in CMAKE, you want to set the CMAKE_INSTALL_PREFIX to the INSTALL dir you created in your Visual Studio Project directory
- in CMAKE, you want to check the box for ENABLE_LAYOUT
- in Visual Studio, in addition to building the ALL_BUILD target as shown in the video, you also want to build the INSTALL target )
-
NOTE: If you install a pre-built binary of libSBML then you must compile SBNW with the same version of Visual Studio as used to build libSBML.
-
Clone the latest revision of the master branch to Documents\Visual Studio 2017\Projects\sbnw via git. In the Documents\Visual Studio 2017\Projects\ folder, run
git clone https://github.com/sys-bio/sbnw.git -
Create BUILD and INSTALL directories in Documents\Visual Studio 2017\Projects\ to hold the build and install files that will be created (the names BUILD and INSTALL are suggestions, not requirements)
-
Download and install CMake (compatible with major version 2 or 3).
-
Open CMake and select Documents\Visual Studio 2017\Projects\sbnw as the source directory.
-
Select Documents\Visual Studio 2017\Projects\BUILD as the build directory.
-
Click on the Advanced option, in the top right section of the main CMake page.
-
Click configure & generate via CMake, choosing a generator that matches the required configuration (32-bit x86 is recommended on Windows; on Linux the default generator is sufficient).
-
In CMake, if you don't see LIBSBML_PREFIX on the list, click on the +AddEntry button in the top right corner and add it. Set the
LIBSBML_PREFIXvariable to point to the directory where libSBML is installed/downloaded. -
If libSBML was compiled with compression support,
LIBSBML_EXTRA_LIBSneeds to be set. If it is not on the list, click on the +AddEntry button in the top right corner and add it. SetLIBSBML_EXTRA_LIBSto include libxml2, libbz2, and libz. On Linux and Mac, this isLIBSBML_EXTRA_LIBS='xml2;bz2;z'. On Windows, you must specify the full path to each dependency in the libSBML third party install folder,
e.g. `LIBSBML_EXTRA_LIBS="\path\to\Documents\Visual Studio 2017\Projects\libSBML-5.17.0-Source\INSTALL\lib\libbz2.lib; path\to\Documents\Visual Studio 2017\Projects\libSBML-5.17.0-Source\INSTALL\lib\libiconv.lib; path\to\Documents\Visual Studio 2017\Projects\libSBML-5.17.0-Source\INSTALL\lib\libxml2.lib; path\to\Documents\Visual Studio 2017\Projects\libSBML-5.17.0-Source\INSTALL\lib\zdll.lib; ws2_32.lib"'. Click configure & generate again. -
If you are compiling with Python support, set
PYTHON_EXECUTABLEto your Python executable,PYTHON_LIBRARYto your Python library, andPYTHON_INCLUDE_DIRto the Python include directory. UseSBNW_LINK_TO_STATIC_LIBSBML=ONif compiling with Python. -
NOTE: In order to statically link to libSBML, specify
SBNW_LINK_TO_STATIC_LIBSBML=ON. Otherwise, the libSBML DLLs must be in the PATH to run any compiled code. -
On Windows, open the generated .sln in Visual Studio, and change the configuration to "Release"; on Linux/Mac simply run make -j4 install from the build directory.
-
(This step was previously used to instruct the user to set the MSVC runtime library. It is now set automatically through CMake. This placeholder serves as a reminder in case this solution breaks at some point.)
-
Windows Specific: In Visual Studio, right click on the INSTALL target and select build. SBNW will be installed to the location stored in CMAKE_INSTALL_PREFIX (ensure your user has write access).
The network viewer is available as a plugin for a pre-release of Tellurium. Simply switch to the network viewer tab, open a model from the test cases (e.g. BorisEJB.xml), and enter the following code:
# The nwed module is used to communicate with the plugin
import nwed
# Get the current model
sbmlstr = nwed.getsbml()
# Print the SBML
print(sbmlstr)
# Load the SBML back into the network viewer
nwed.setsbml(sbmlstr)
The C API is exposed via graphfab/autolayoutc_api.h. There is an example in sandbox/basic that demonstrates all the functionality needed to get the layout working:
// type to store layout info
gf_layoutInfo* l;
// load the model
gf_SBMLModel* mod = gf_loadSBMLbuf(buf);
// options for layout algo
fr_options opt;
// read layout info from the model
l = gf_processLayout(mod);
// randomize node positions
gf_randomizeLayout(l);
// do layout algo
opt.k = 20.;
opt.boundary = 1;
opt.mag = 0;
opt.grav = 0.;
opt.baryx = opt.baryy = 500.;
opt.autobary = 1;
gf_doLayoutAlgorithm(opt, l);
// save layout information to new SBML file
gf_writeSBMLwithLayout(outfile, mod, l);
// run destructors on the model
gf_freeSBMLModel(mod);
// run destructors on the layout
gf_freeLayoutInfo(l);
This project is licensed under the BSD 3-clause license:
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
- Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
- Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
- Neither the name of The University of Washington nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDERS OR ANYONE DISTRIBUTING THE SOFTWARE BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
This work was funded by the generous support of the National Institute of General Medical Sciences of the National Institutes of Health under award R01-GM081070