Analysis and synthesis tools for OCCAM hPF simulations
Project description
OccamTools
·





Analysis and synthesis tools for OCCAM molecular dynamics/hybrid particle-field simulations.
Input to OCCAM consists mainly of three file types; fort.1 (simulation metadata), fort.3 (particle and bond parameters), and fort.5 (positions and bond structure). The output from the run is a fort.8 file, adhering to the .xyz file format. The occamtools python package provides a reader for these file formats (both input and output) and generates a single object containing all the information about the simulation, making analysis of simulation runs and comparison between runs easier.
Installation
Install by (requires python >= 3.6)
> pip install occamtools
Usage
Loading data · Loading simulation data is done by
import numpy as np
from occamtools import OccamData
data = OccamData('your/file/or/directory/here')
where the data object now holds all information about the simulation run, e.g. print what kinds of particles a simulation consists of
print('Simulation consists of...')
for type_name in data.type_dict:
num_type = sum(data.type == data.type_dict[type_name])
print(f' - {num_type} particles of type {type_name}')
# Simulation consists of...
# - 400 particles of type H
# - 100 particles of type C
# - 250 particles of type Ar
# ...
or make a simple plot of particles in a section of the simulation box diffusing (requires pip install asciichartpy)
from asciichartpy import plot
indices = (data.x[0, :] > 20.0) & (data.x[0, :] < 25.0)
diffused = data.x[-1, indices]
bins, hist = np.histogram(diffused)
print(plot(bins.tolist(), hist.tolist()))
# 9.00 ┤ ╭─╮
# 8.00 ┤ │ ╰╮
# 7.00 ┤ │ │
# 6.00 ┤ ╭╯ ╰╮
# 5.00 ┤ │ ╰╮
# 4.00 ┤ │ │
# 3.00 ┤╭╯ │
# 2.00 ┤│ ╰╮
# 1.00 ┼╯ ╰
or plot the deviations of the total kinetic energy from the mean over the simulation run (again requires pip install asciichartpy)
kinetic_energy_deviations = data.kinetic_energy - np.mean(data.kinetic_energy)
print(plot(kinetic_energy_deviations.tolist()))
# 60.53 ┤ ╭╮
# 50.62 ┤ ││ ╭╮╭╮
# 40.71 ┤ ││ ││││ ╭╮
# 30.81 ┤ ││ ││││ ││ ╭╮ ╭─
# 20.90 ┤ ╭╮ ││ ││││ ╭─╮ ││ ││ │
# 10.99 ┤ ││ ╭─╮ ╭╮ ╭╯│ ││││╭╮ │ │ ╭╮ ││ ││ │
# 10.09 ┤ ││ │ │ ││ │ │ │││││╰╮ │ ╰╮││ ││ ╭╮ ││╭╮╭╮│
# 0.18 ┼╮││ │ │ ││ │ ╰╮ │││╰╯ │ │ ╰╯│╭╮ │╰╮││╭╮│││││││
# -00.73 ┤│││ │ │ ││ │ ╰─╯││ │╭╯ ││╰──╯ ╰╯││││││││││
# -10.64 ┤│││╭╯ │ ││╭╯ ││ ││ ╰╯ ││││││││╰╯
# -20.54 ┤││╰╯ │ │╰╯ ╰╯ ╰╯ ││││││╰╯
# -30.45 ┤╰╯ ╰─╯ ││││╰╯
# -40.36 ┤ ││││
# -50.26 ┤ ││╰╯
# -60.17 ┤ ╰╯
File storage
·
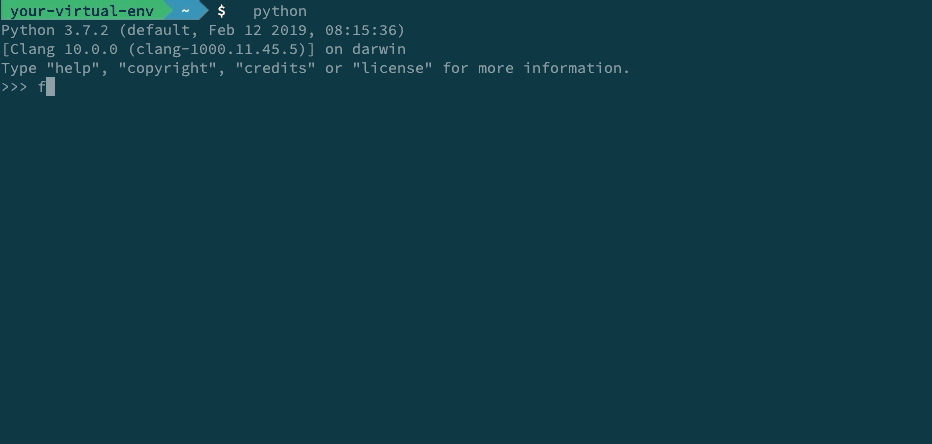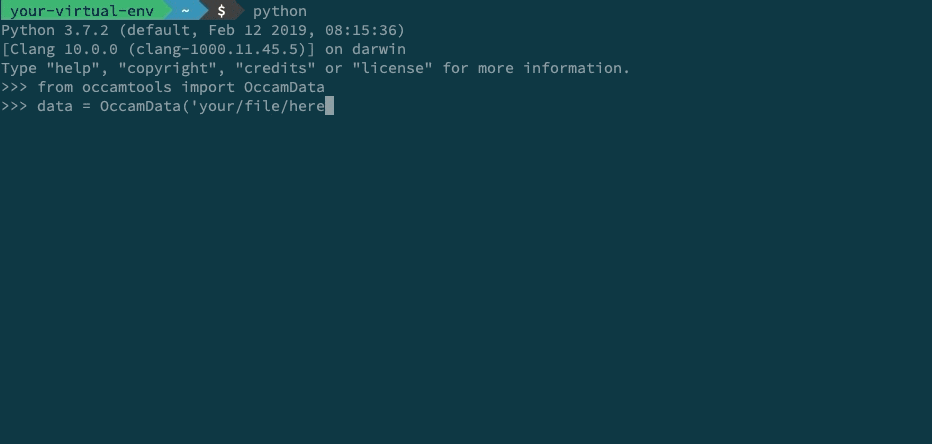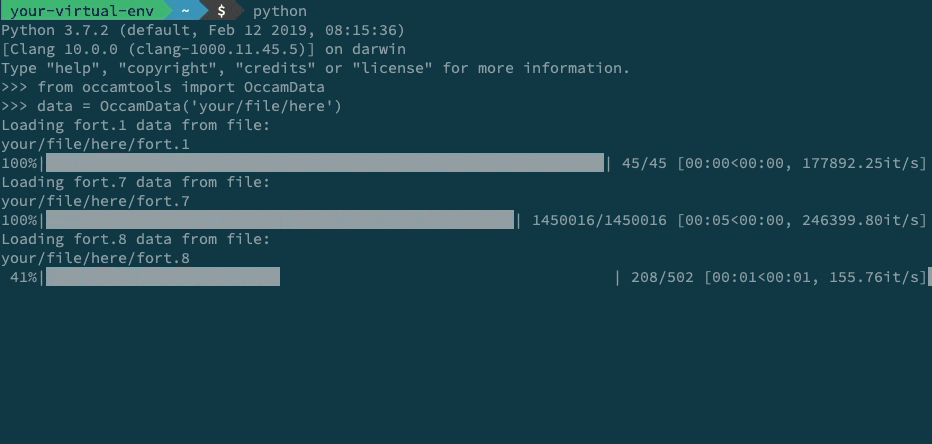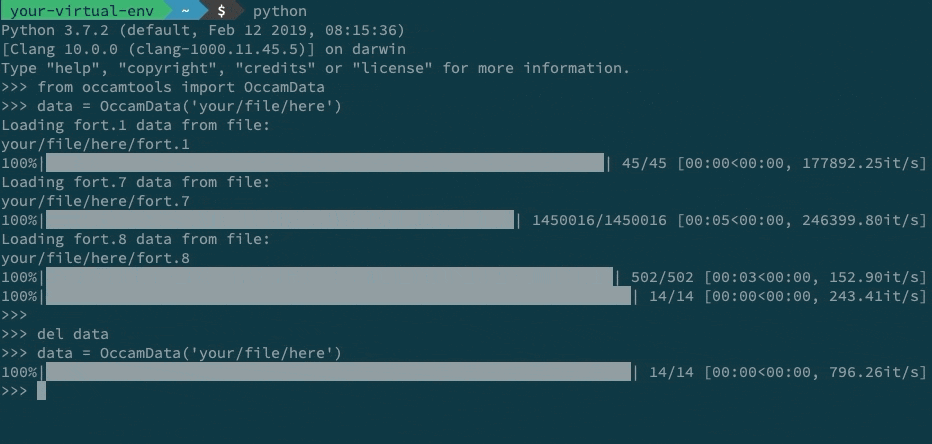
Behind the scenes, .npy (for numpy arrays) and .json (for anything else) files are used to represent the simulation data. By default, loading a simulation run causes the saving of small (relative to the original fort.5/7/8) binary files containing the data. These are used to load from on subsequent calls. This means calls to OccamData.load('your/file/here') of OccamData('your/file/here') will be significantly faster after the first call. In this specific example, a 25x speedup is achieved (but your mileage may vary).
Running tests
Inside the occamtools directory, do
> pip3 install pytest
> pytest -v
OCCAM
OCCAM is a program for Molecular Dynamics Simulations able to perform Hybrid Particle-Field (PF) Theoretical Molecular Dynamics simulations. This recent PF technique combines molecular dynamics (MD) and self consistent field theory (SCF). Read more.
Changelog
0.3.4: Add bond energy and angular bond energy to Fort7 reader.
0.3.3: Add option to not save .npy files when loading OccamData objects. Fix a bug causing errors when reading very short .xyz files. Fix a bug causing the grid size to not correctly update when using replace_in_fort3.
0.3.2: Add testing with python alpha version 3.8-dev, stream line travis integration and coverage reporting.
0.3.1: Move the bins keyword argument to histogram from an explicit argument to **kwargs handled by np.histogram.
0.3.0: Add histogram computation capabilities.
0.2.7: Add the velocity_traj flag to fort1 file reader.
0.2.6: Add proper testing for python 3.6 and 3.7 using tox.
0.2.5: Change python version required to >=3.6 (from >=3.7).
0.2.4: Add functionality for reading .xyz files with additional velocity information, as output by OCCAM when the velocity_traj flag is set in fort.1.
0.2.3: Code clean-up.
0.2.2: Extend repace_in_fort3 to allow for changing compressibility and non-bonded interactions. Fix a bug causing new particle types added to break the chi matrix when writing fort.3 files.
0.2.1: Update the __all__ variable of __init__.py to reflect newly added classes and methods.
0.2.0: Add functionality for editing fort.3 files (in-place or creating new ones).
0.1.0: Add functionality for editing fort.1 files (in-place or creating new ones).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.