Grep-like tool for retrieving matching sequence records from a FASTA file
Project description
Better FASTA Grep

Better FASTA Grep, or BFG for short, is a Grep-like utility for retrieving matching sequence records from a FASTA file. Given one or more patterns and a FASTA file, it searches the file for matching headers and or sequences and outputs any matching headers, sequences, or both.
Features
- Search headers, sequences, or both
- Search via regular expressions or plain strings
- Case-insensitive search
- Select non-matching sequence records
- Count the number of matches
- Display line numbers in the result
- Sequence records, not individual lines, are selected
- Multi-line sequences are treated as singular units
- Flexible output options: output headers, sequences, or both
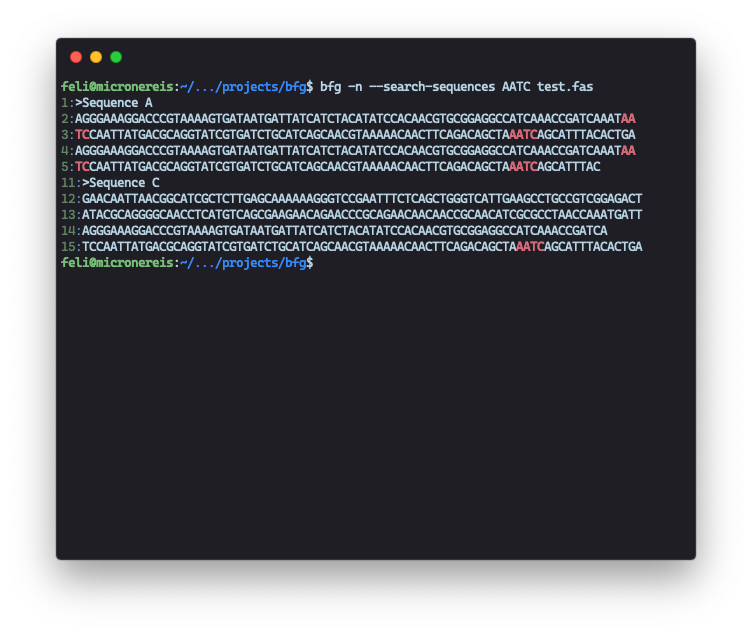
A screenshot of running BFG under macOS Mojave.
Quick installation
The easiest way to install this program is via pip:
pip install better_fasta_grep
You can then launch bfg using one of the following commands:
bfg --help
better_fasta_grep --help # equivalent
Documentation
© Department for Animal Evolution and Biodiversity 2019
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
better_fasta_grep-1.0.4.tar.gz
(148.8 kB
view hashes)












