Relational Graph Attention Network for Spatial Transcriptome Analysis
Project description
RGAST
RGAST: Relational Graph Attention Network for Spatial Transcriptome Analysis [paper]
This document will help you easily go through the RGAST model.
Installation
To install our package, run
pip install RGAST
You can also clone the repo and install it in editable mode:
git clone https://github.com/GYQ-form/RGAST.git
cd RGAST
pip install -e .
Usage
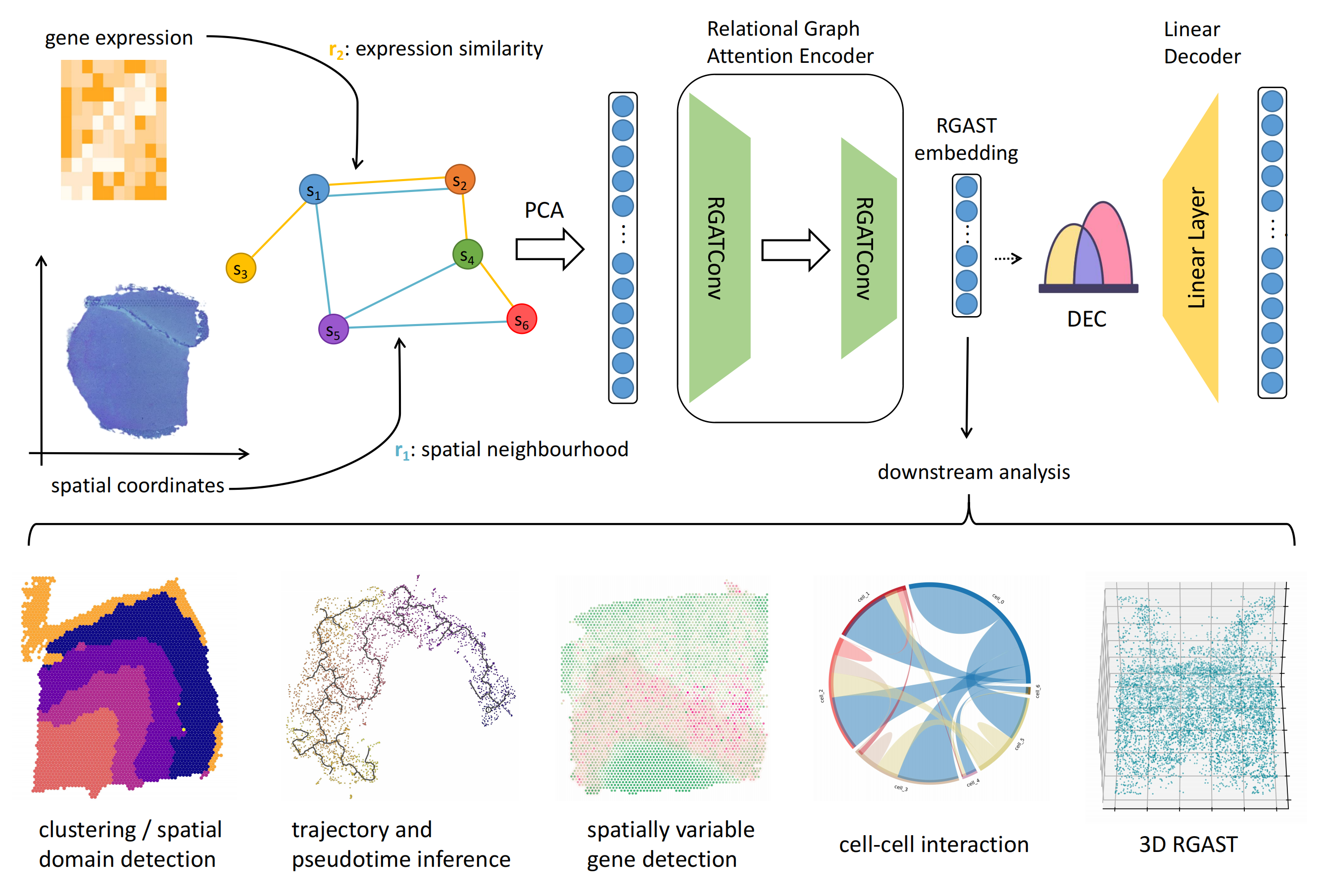
RGAST (Relational Graph Attention network for Spatial Transcriptome analysis) constructs a relational graph attention network to learn the representation of each spot in the spatial transcriptome data. Plus the attention mechanism, RGAST considers both gene expression similarity and spatial neighbor relationships in constructing the graph network, enabling a more comprehensive and flexible representation of the spatial transcriptome data. RGAST can be used in many ST analysis:
- spatial domain identification
- cell trajectory inference
- spatially variable gene (SVG) detection
- uncover spatially resolved cell-cell interactions
- reveal intricate 3D spatial patterns across multiple sections of ST data
Tutorial
We have prepared several basic tutorials in https://github.com/GYQ-form/RGAST/tree/main/tutorial. You can quickly hands on RGAST by going through these tutorials. Model parameters trained in our study are also released in https://github.com/GYQ-form/RGAST/tree/main/model_path.